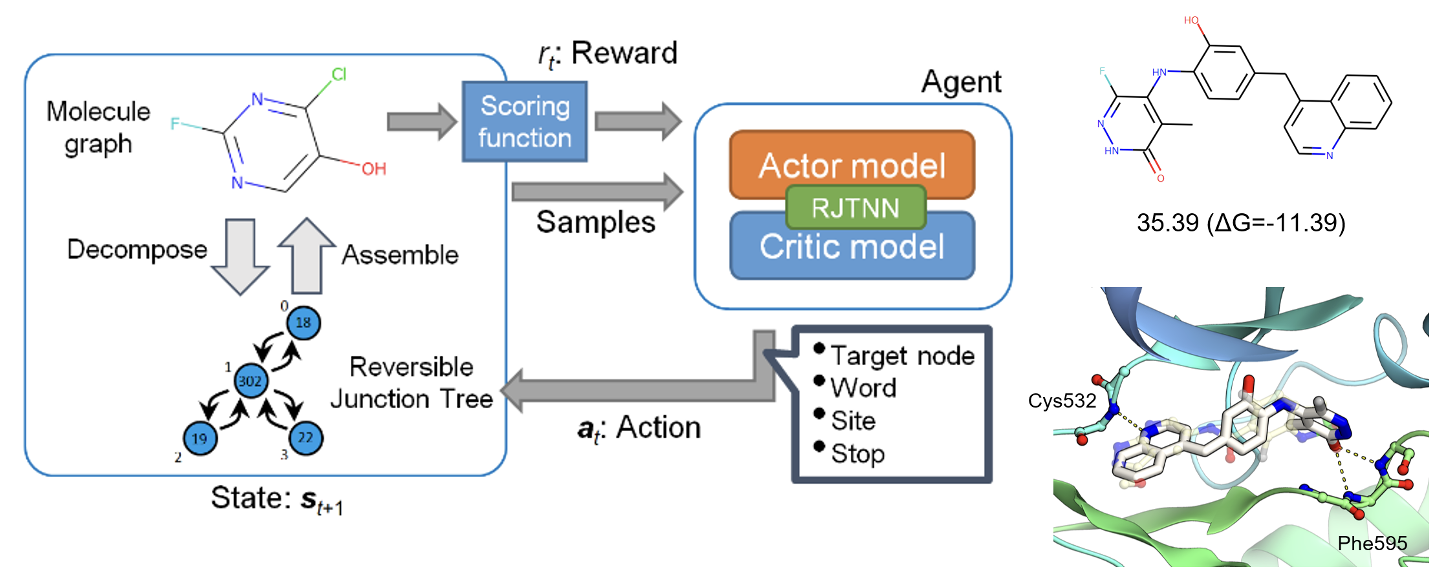
Research
Our department combines computational physical chemistry with informatics to understand biological systems. We use molecular simulations and quantum calculations alongside machine learning and structural informatics to design biomolecules for drug discovery applications.
Research Topics
Dynamics of Biological Macromolecules by Molecular Dynamics Simulation
- Elucidation of GPCR dynamics and signaling mechanisms by MD simulation Verification of LPA6 receptor lateral access mechanism
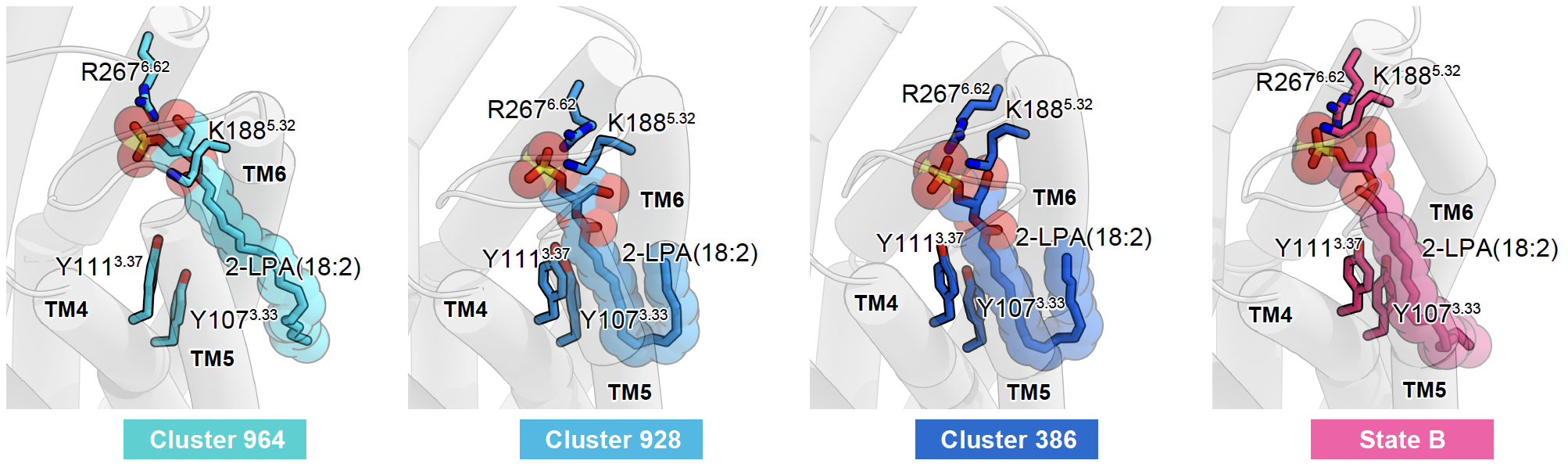
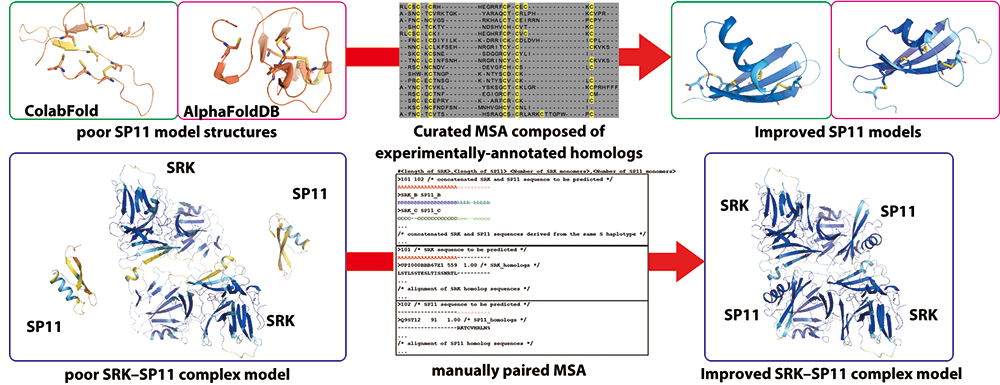
Protein structure prediction using AlphaFold and application of the structural information
- Haplotype-specific SRK-SP11 complexes controlling self-incompatibility in Brassica oleracea were predicted with high confidence using multiple sequence alignment (MSA) and ColabFold. SRK-SP11 complex that regulates self-incompatibility in Brassica rapa with a high degree of confidence. Comprehensive computational analysis of the SRK–SP11 molecular interaction underlying self-incompatibility in Brassicaceae using improved structure prediction for cysteine-rich proteins
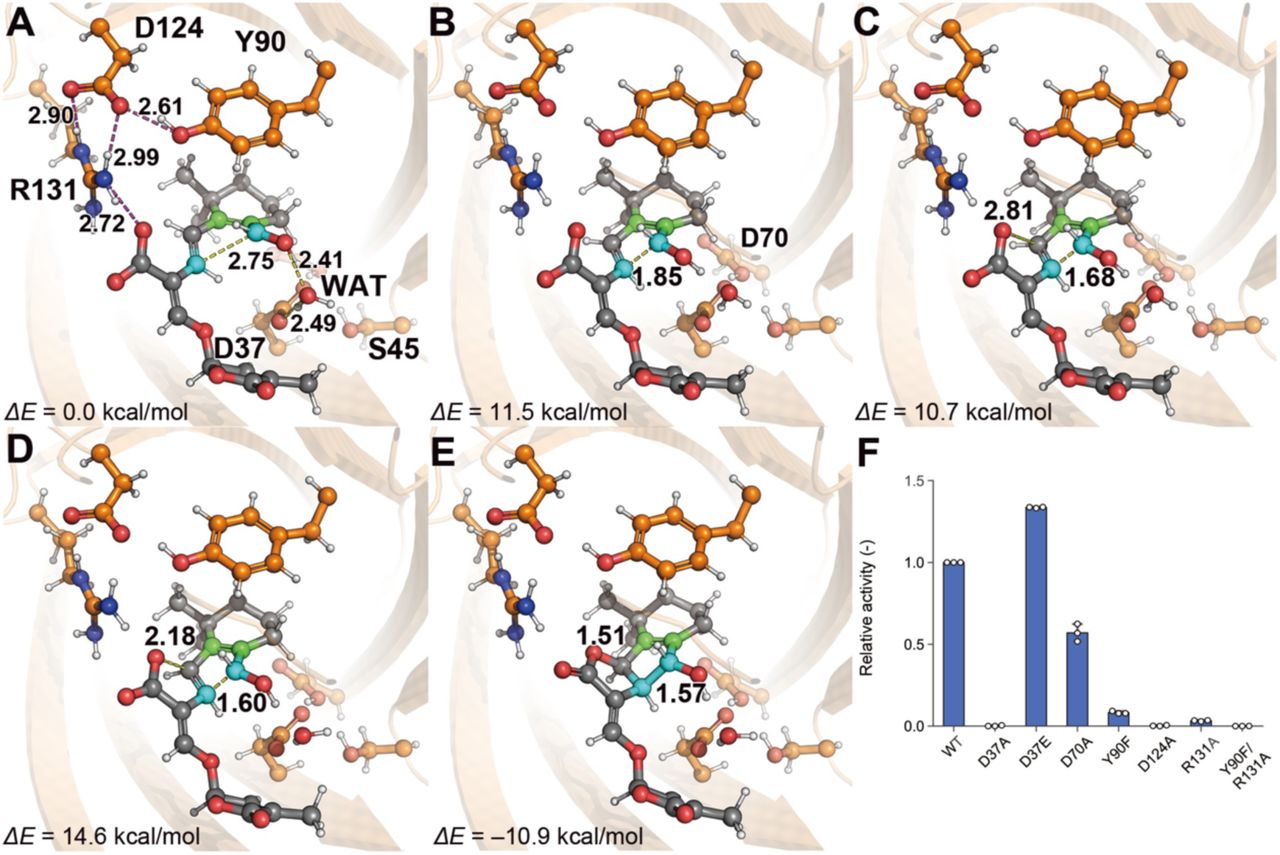
Elucidation of the reaction mechanism of enzymes using predicted protein structures and QM/MM simulation
-
Predicted structure of AlphaFold2 combined with MD simulation and QM/MM(ONIOM) method to elucidate the reaction mechanism of the enzyme Insights into stereoselective ring formation in canonical strigolactone: Discovery of a dirigent domain-containing enzyme catalyzing orobanchol synthesis
-
QM/MM simulation for p-Hydroxybenzoate hydroxylase (PHBH)
Development of a method for analyzing the dynamics of biological macromolecules by combining molecular simulation and deep learning
- Applications of Deep Learning to Structural Biology
- Protein Property Prediction Using Graph Neural Networks
- Application of Deep Learning to Structure-Based Small and Medium Molecular Drug Discovery